|
Yichun He I am an Eric and Wendy Schmidt Fellow at the Broad Institute of MIT and Harvard.
I received my Ph.D in the School of Enginnering and Applied Sciences at Harvard University,
and bachelor's degree in Electrical Engineering at the University of Science and Technology of China with the highest honor Guomoruo Scholarship.
|

|
ResearchI am passionate about accelerating discovery in biology and health through advanced AI/ML approaches. My work spans multiple directions, including:
# Co-first authors; * Corresponding authors. |
(1) Characterize whole-brain spatially resolved cell types at molecular resolution with scalable AI and spatial transcriptomics |
|
|
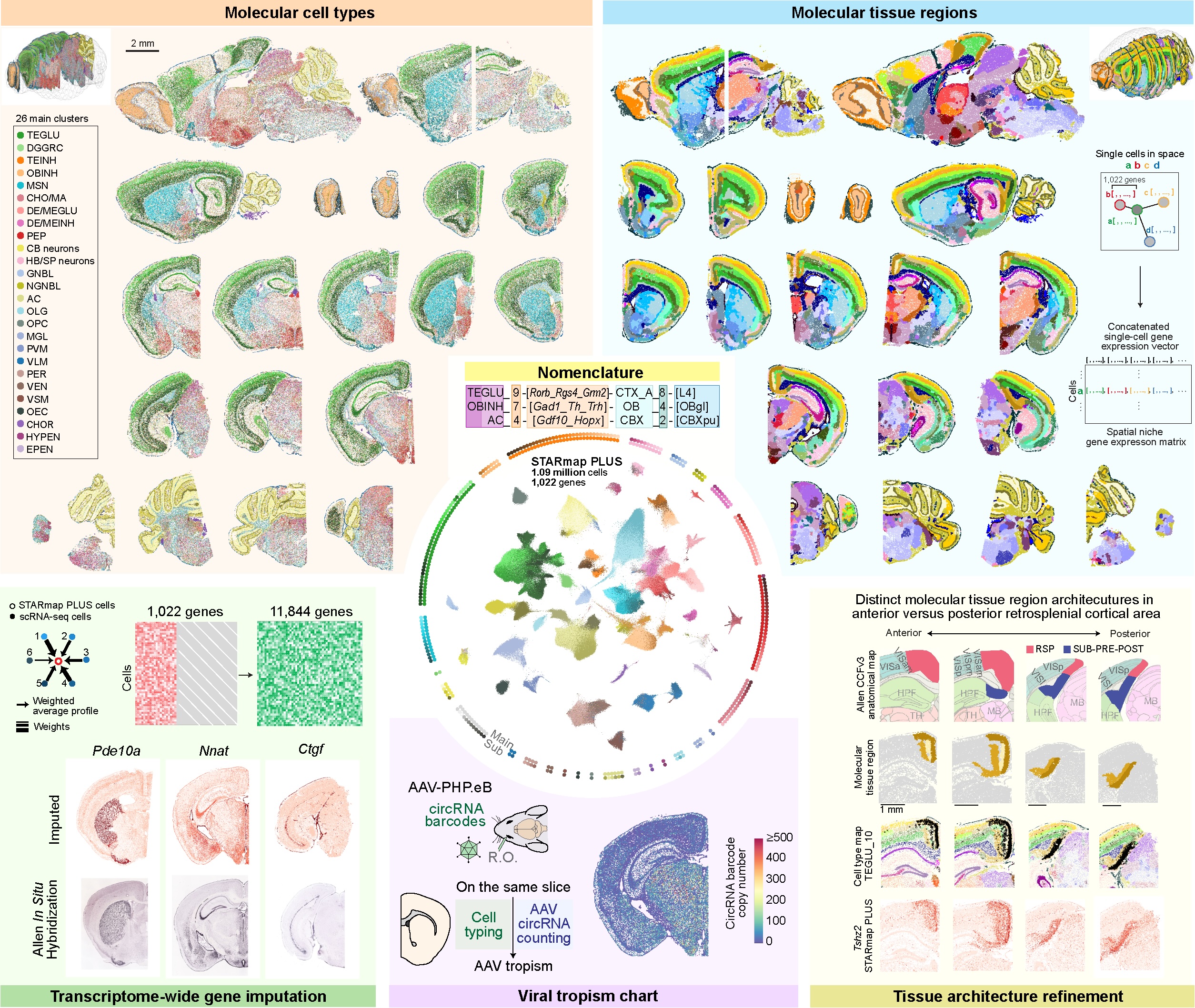
Spatial Atlas of the Mouse Central Nervous System at Molecular Resolution
Hailing Shi#, Yichun He#, Yiming Zhou#, Jiahao Huang, Kamal Maher, Brandon Wang, Zefang Tang, Shuchen Luo, Peng Tan, Morgan Wu, Zuwan Lin, Jingyi Ren, Yaman Thapa, Xin Tang, Ken Y. Chan, Benjamin E. Deverman, Hao Shen, Albert Liu, Jia Liu* & Xiao Wang* Nature, 2023 Github Cover and Perspective highlight in Nature as part of collection for whole mouse brain spatial atlas, "BRAIN Initiative Cell Census Network 2.0 – Whole Mouse Brain". Google Map of the Mouse Brain at Molecular Resolution. |
|
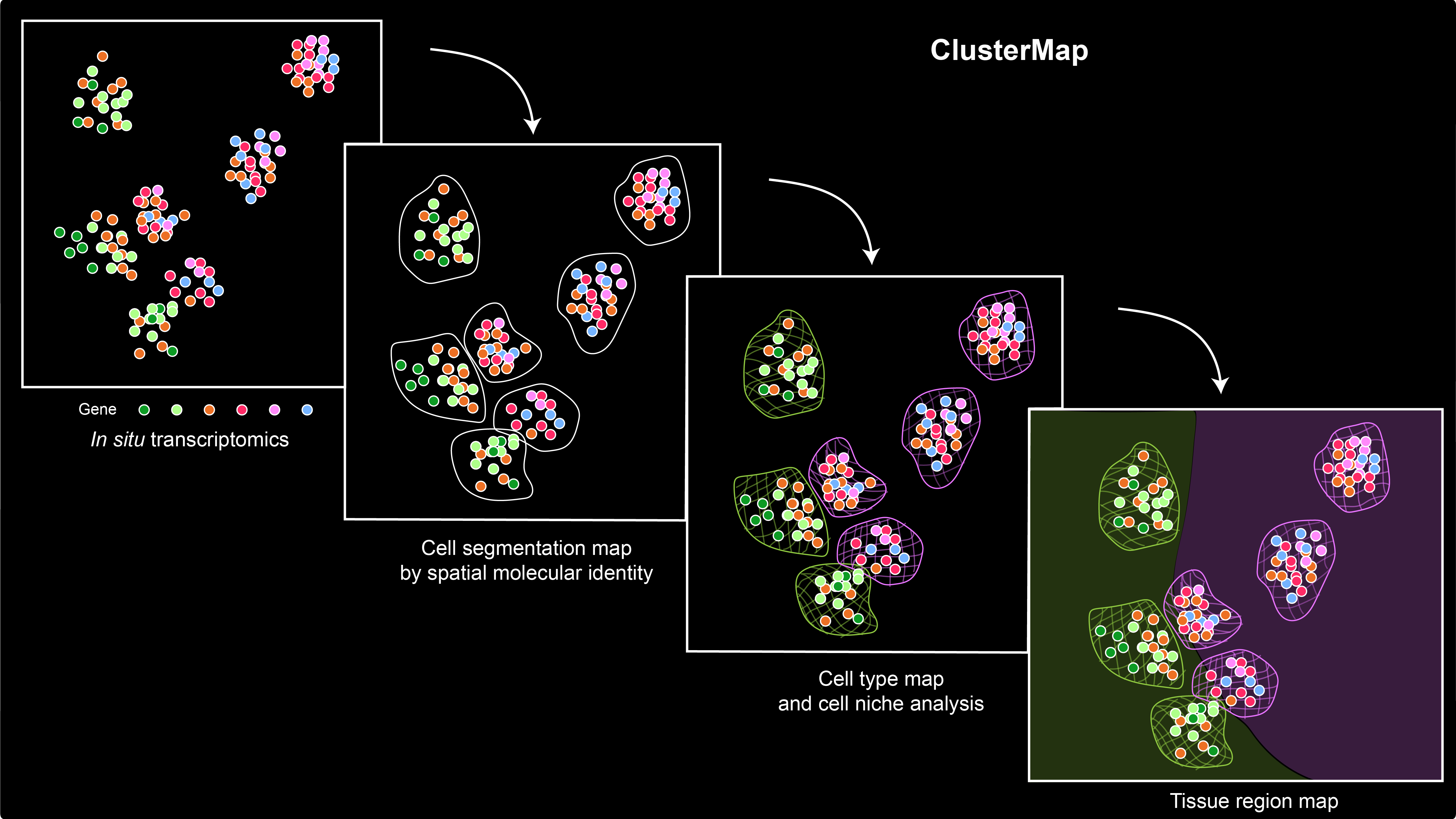
ClusterMap for multi-scale clustering analysis of spatial gene expression
Yichun He#, Xin Tang#, Jiahao Huang, Jingyi Ren, Haowen Zhou, Kevin Chen, Albert Liu, Hailing Shi, Zuwan Lin, Qiang Li, Abhishek Aditham, Johain Ounadjela, Emanuelle I. Grody, Jian Shu, Jia Liu* & Xiao Wang* Nature Communications, 2021 ClusterMap package Research develops new way to map RNAs in the cell. |
|
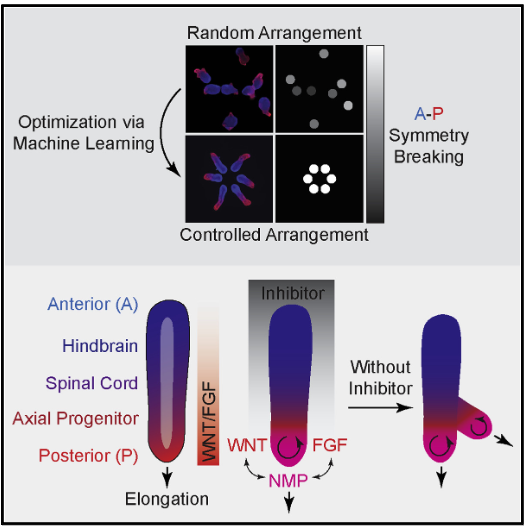
Controlling organoid symmetry breaking uncovers an excitable system underlying human axial elongation
Giridhar M. Anand, Heitor C. Megale, Sean H. Murphy, Theresa Weis, Zuwan Lin, Yichun He, Xiao Wang, Jia Liu, Sharad Ramanathan* Cell, 2023 |
(2) Bridge cellular features with spatial contexts across various technologies with contextual AI |
|
|
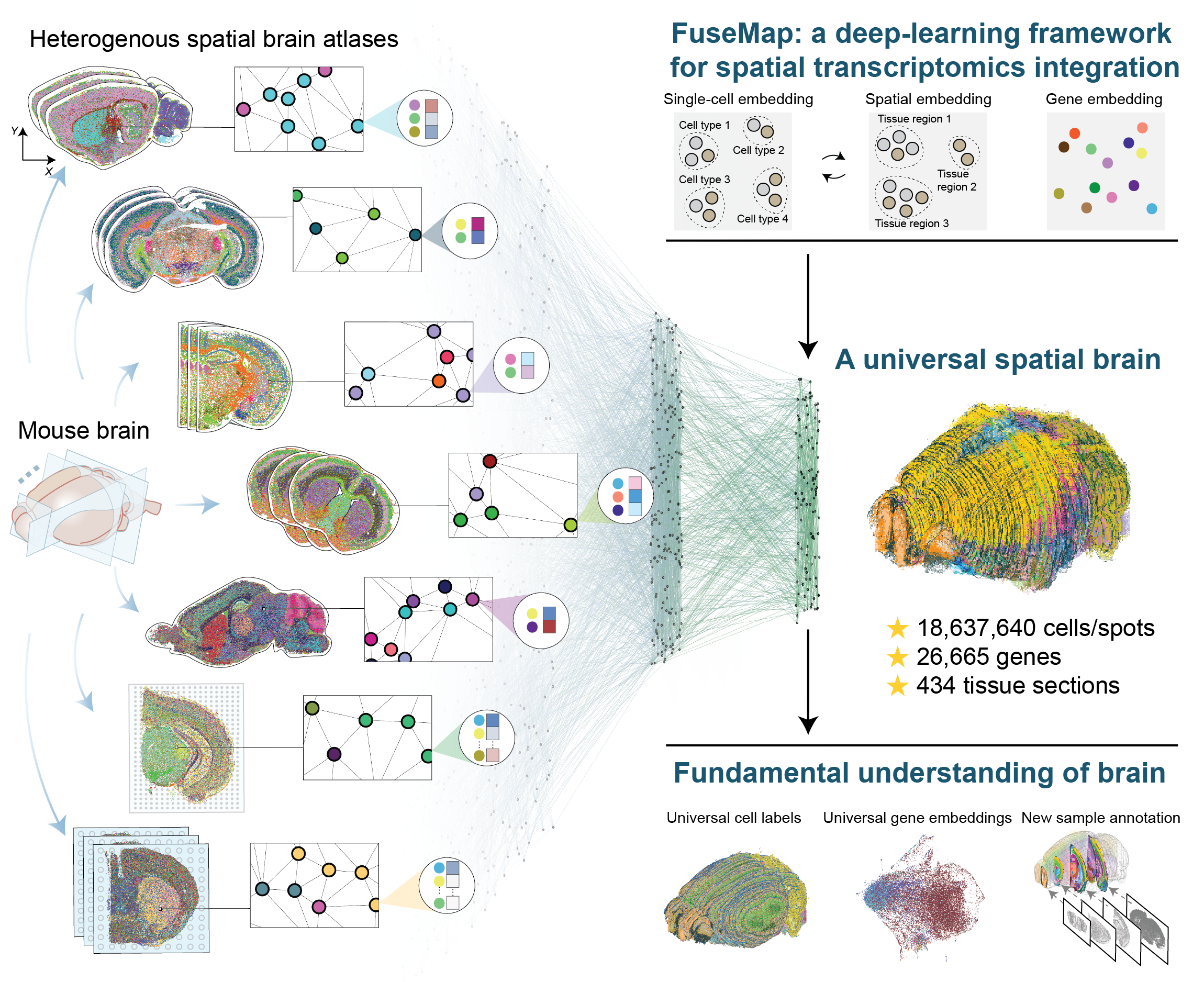
Towards a universal spatial molecular atlas of the mouse brain
Yichun He, Hao Sheng, Hailing Shi, Wendy Xueyi Wang, Zefang Tang, Jia Liu*, and Xiao Wang* bioRxiv, 2024 Data portal of the molecular common coordinate framework (molCCF) FuseMap package |
|
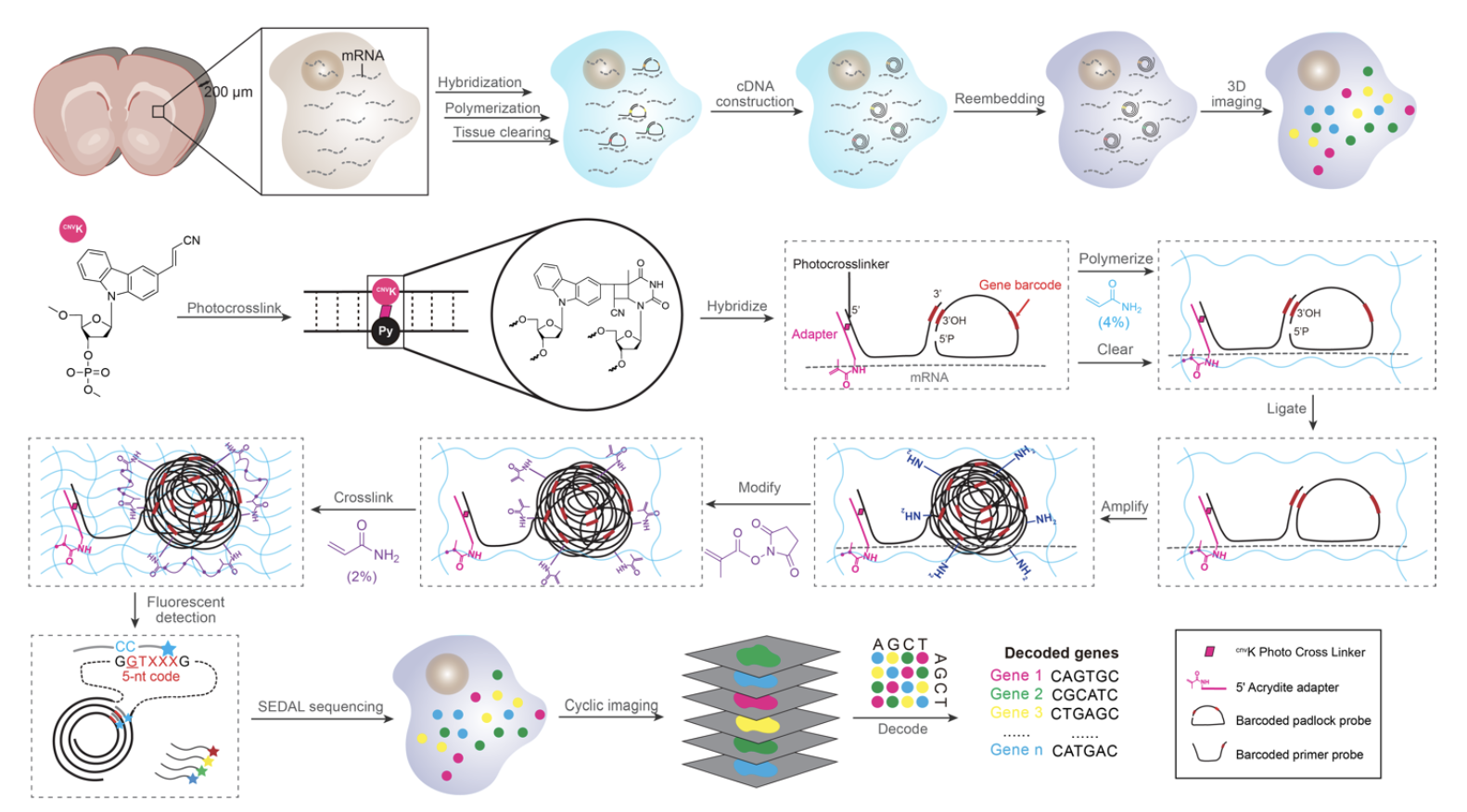
Scalable spatial single-cell transcriptomics and translatomics in 3D thick tissue blocks
Xin Sui#, Jennifer A. Lo#, Shuchen Luo, Yichun He, Zefang Tang, Zuwan Lin,Yiming Zhou, Wendy Xueyi Wang, Jia Liu, Xiao Wang* bioRxiv, 2024 |
|
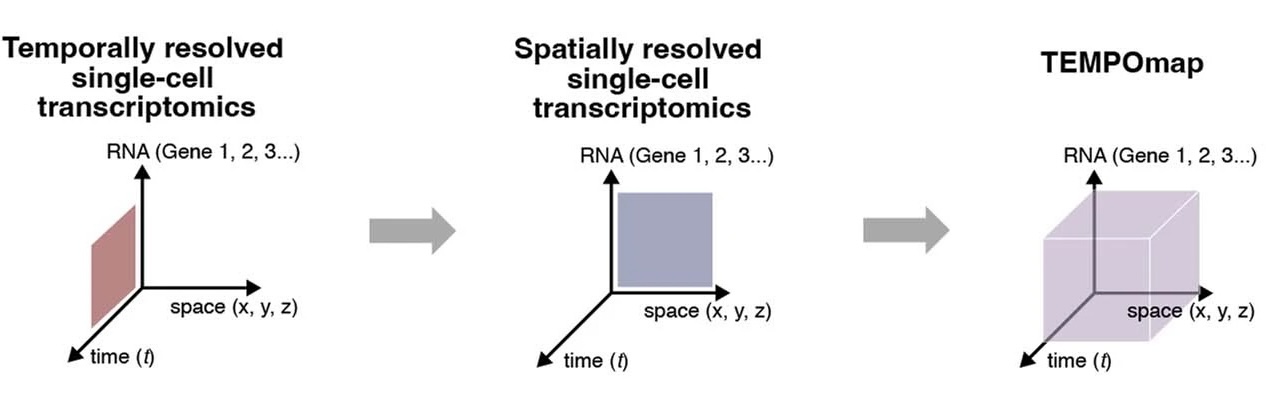
Spatiotemporally resolved transcriptomics reveals the subcellular RNA kinetic landscape
Jingyi Ren#, Haowen Zhou#, Hu Zeng#, Connie Kangni Wang, Jiahao Huang, Xiaojie Qiu, Xin Sui, Qiang Li, Xunwei Wu, Zuwan Lin, Jennifer A. Lo, Kamal Maher, Yichun He, Xin Tang, Judson Lam, Hongyu Chen, Brian Li, David E. Fisher, Jia Liu & Xiao Wang* Nature Methods, 2023 |
(3) Decode dynamic biological processes in long term with real-time AI and brain computer interfaces |
|
|
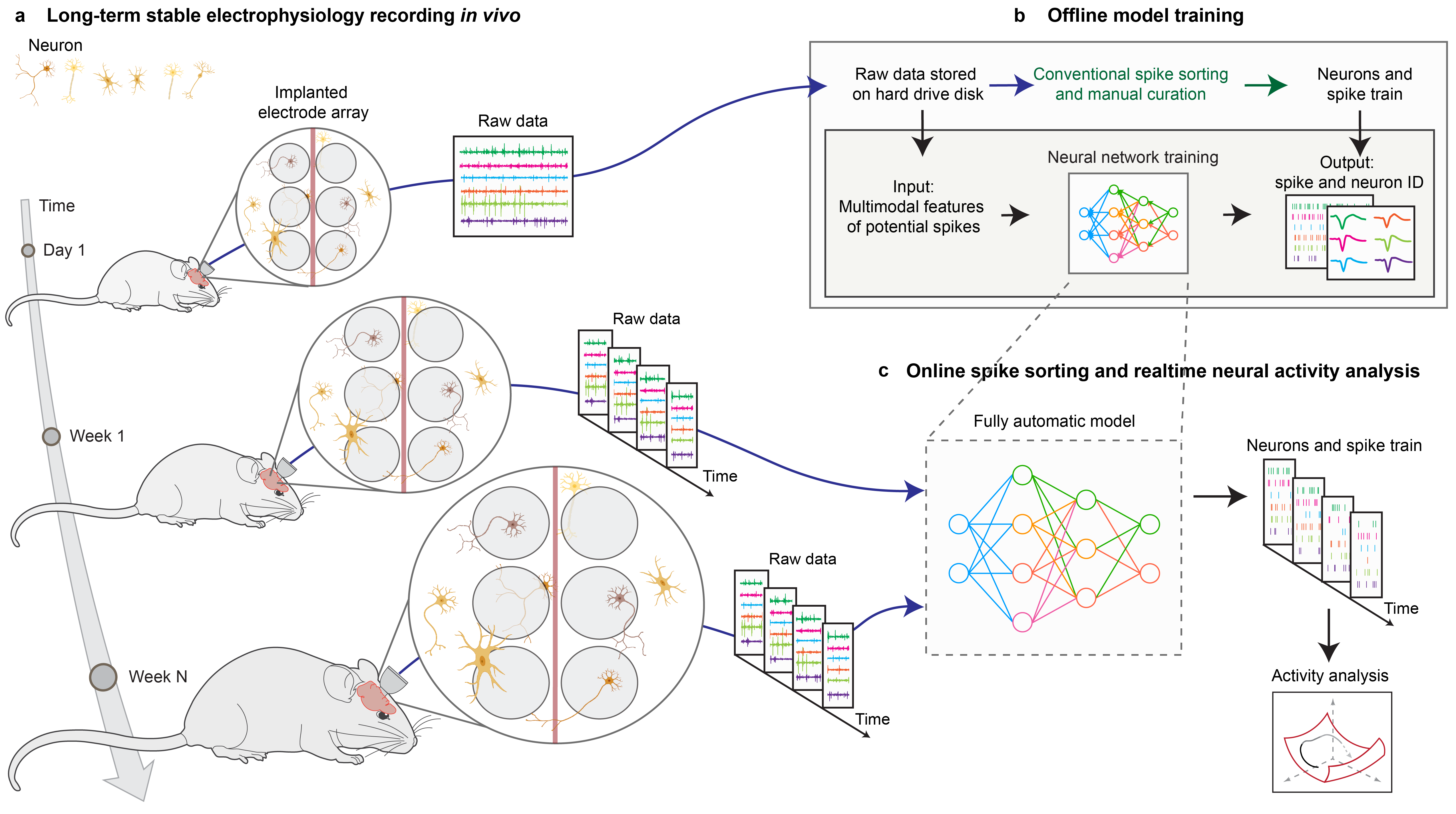
End-to-end multimodal deep learning for real-time decoding of months-long neural activity from the same cells
Yichun He#, Arnau Marin-Llobet#, Hao Sheng, Ren Liu, Jia Liu* bioRxiv, 2024 AutoSort package |
|
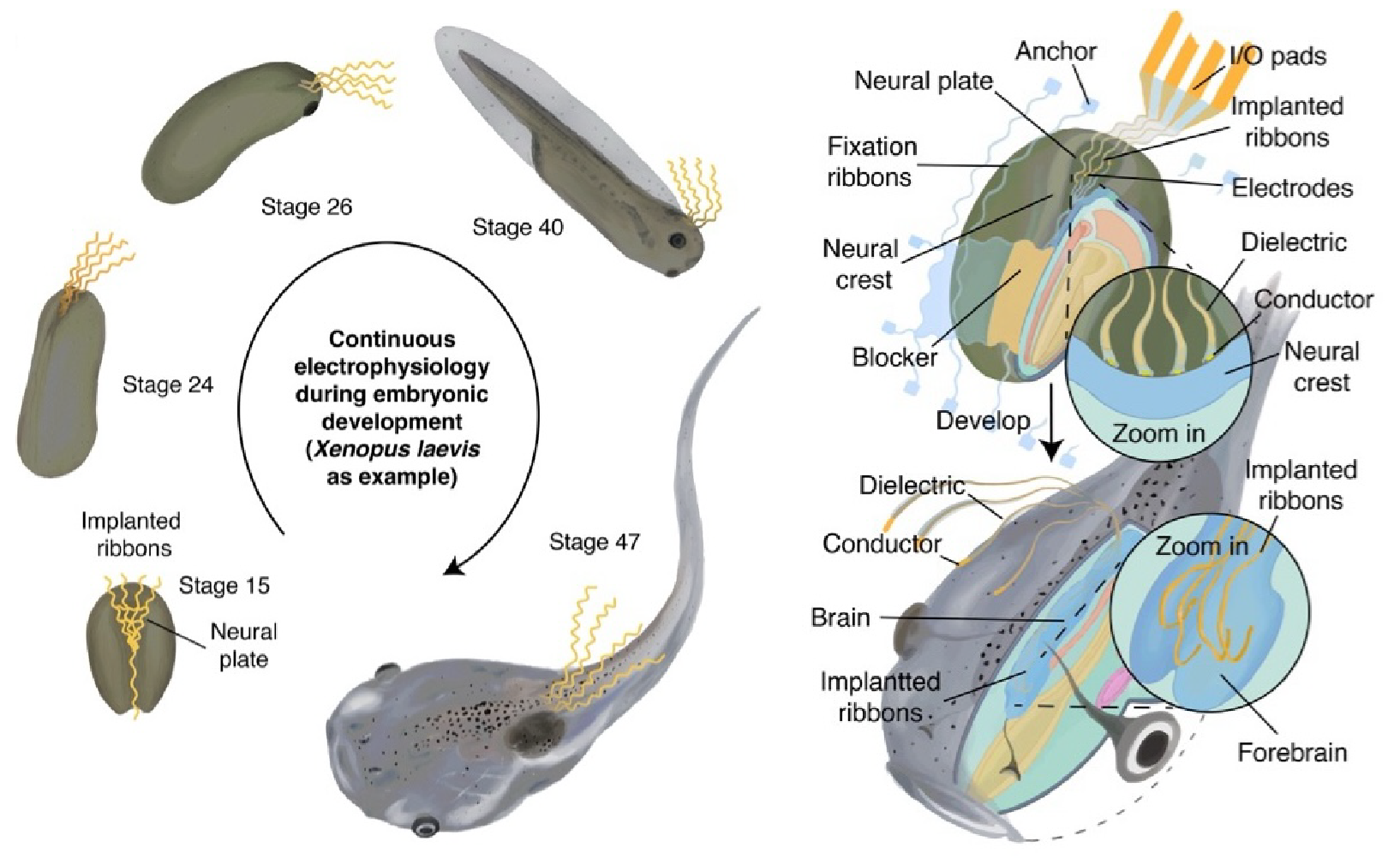
Brain implantation of tissue-level-soft bioelectronics via embryonic development
Hao Sheng#, Ren Liu#, Qiang Li#, Zuwan Lin#, Yichun He, Thomas Blum, Hao Zhao, Xin Tang, Wenbo Wang, Lishuai Jin, Zheliang Wang, Emma Hsiao, Paul Le Floch, Hao Shen, Ariel J. Lee, Rachael Alice Jonas-Closs, James Briggs, Siyi Liu, Daniel Solomon, Xiao Wang, Nanshu Lu, and Jia Liu* Nature, 2025 Cover highlight in Nature |
|
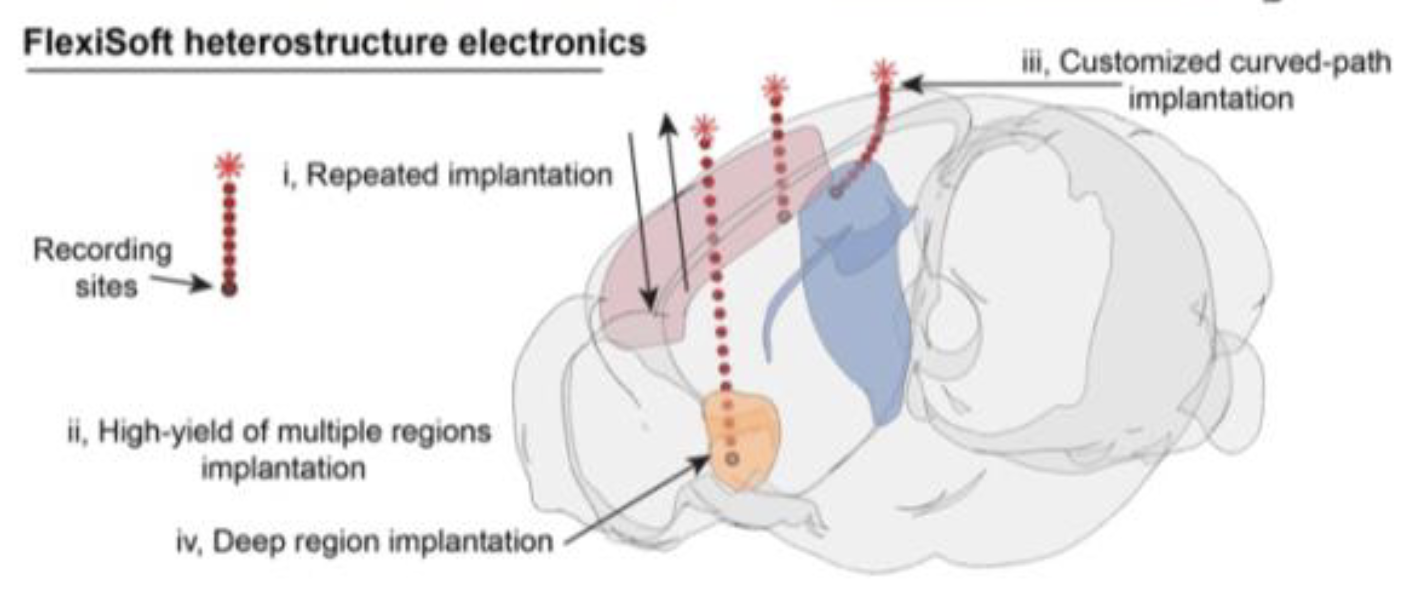
Plastic-elastomer heterostructure for robust flexible brain-computer interfaces
Xinyi Lin, Xinhe Zhang, Zheliang Wang, Juntao Chen, Jaeyong Lee, Ariel J. Lee, Hang Yang, Antoine Remy, Hao Shen, Yichun He, Hao Zhao, Xuyue Zhang, Wenbo Wang, Almir Aljović, Joost J. Vlassak, Nanshu Lu, Jia Liu* bioRxiv, 2025 |
|
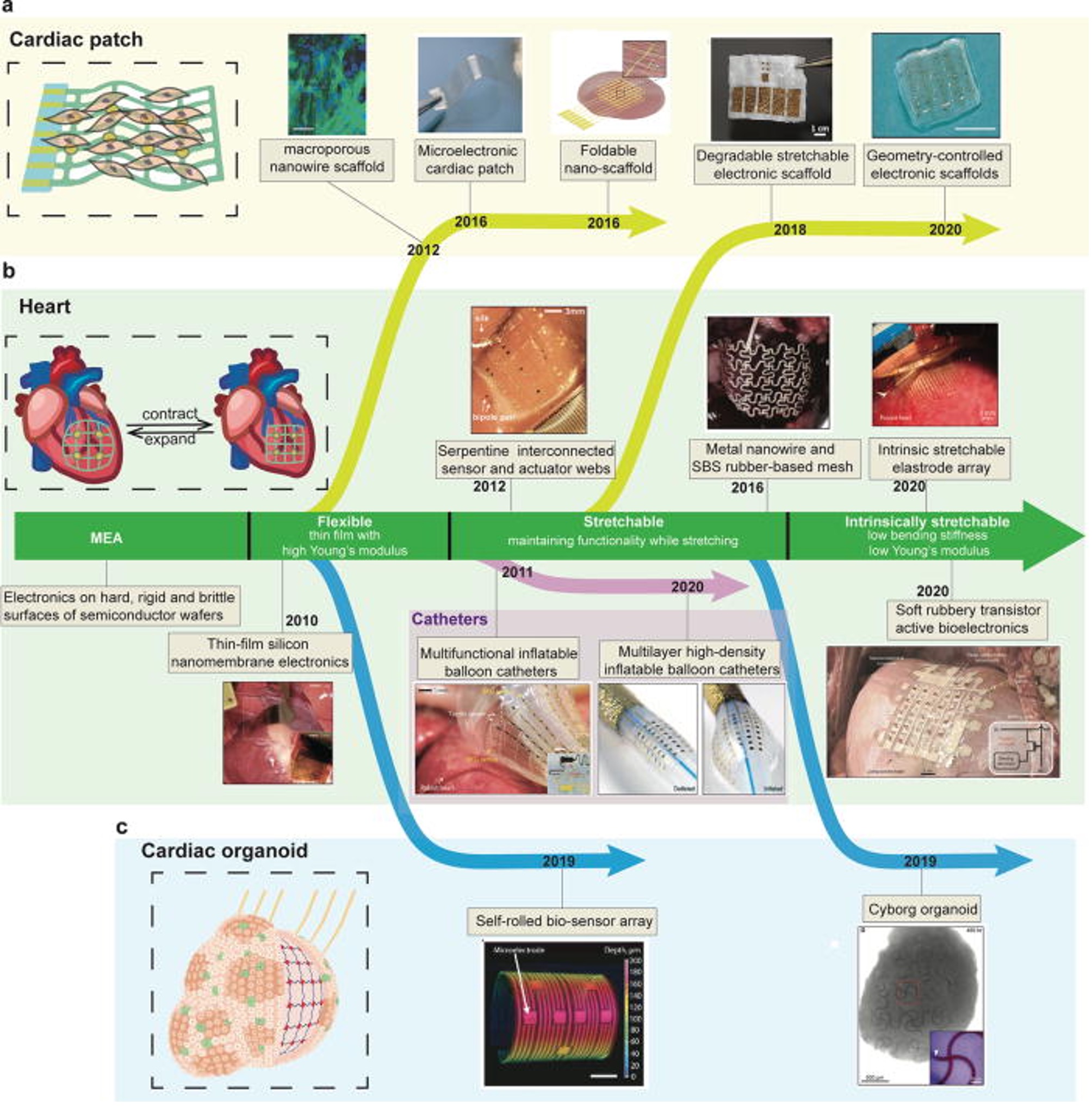
Soft bioelectronics for cardiac interfaces
Xin Tang#, Yichun He# Jia Liu*, Biophysics Reviews, 2022 Editor's highlight |
(4) Integrate diverse biological data with multi-modal AI and single-cell multi-omics |
|
|
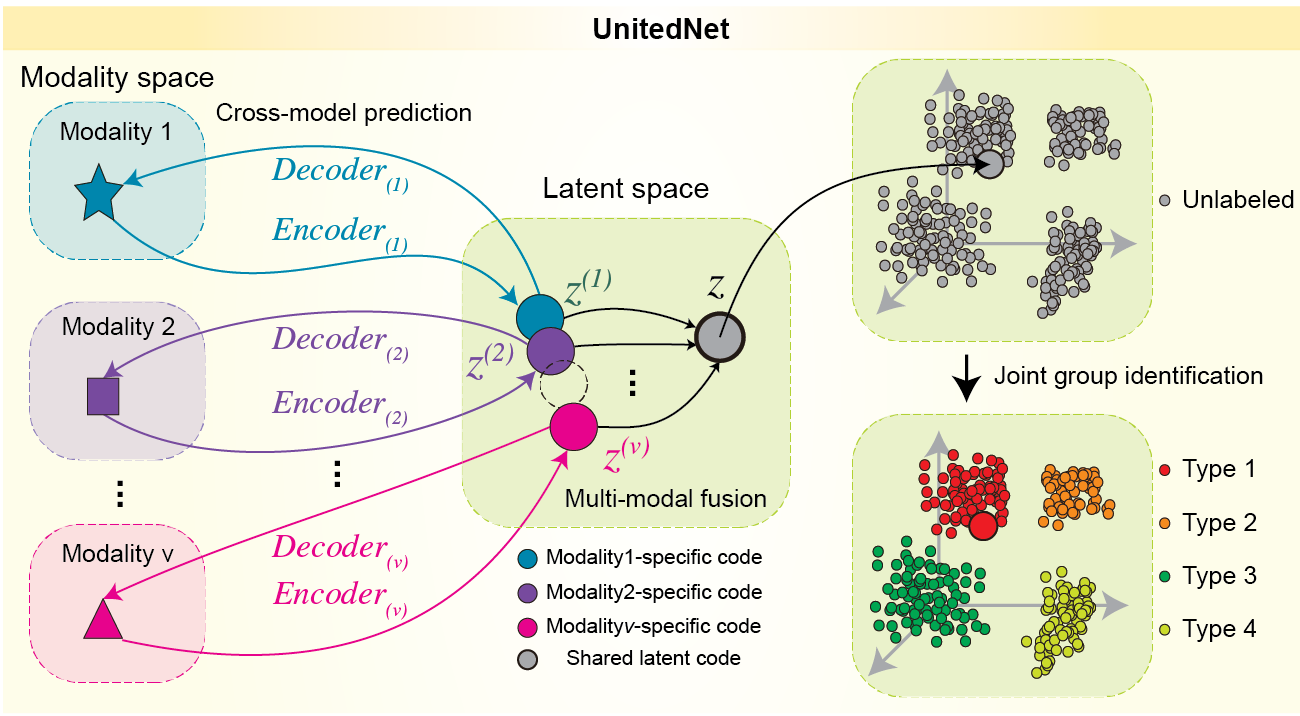
Multi-task learning for single-cell multi-modality biology
Xin Tang#, Jiawei Zhang#, Yichun He#, Xinhe Zhang, Zuwan Lin, Sebastian Partarrieu, Emma Bou Hanna, Zhaolin Ren, Hao Shen, Yuhong Yang, Xiao Wang, Na Li, Jie Ding* & Jia Liu* Nature Communications, 2023 UnitedNet package Editor highlights in Biotechnology and Methods and Computational Science. |
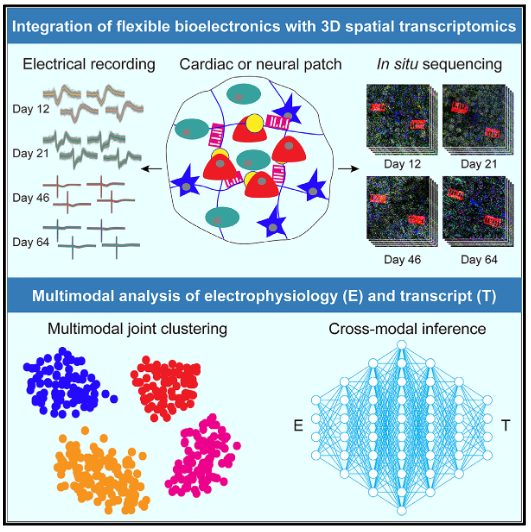
|
Multimodal charting of molecular and functional cell states via in situ electro-seq
Qiang Li#, Zuwan Lin#, Ren Liu#, Xin Tang#, Jiahao Huang, Yichun He, Xin Sui, Weiwen Tian, Hao Shen, Haowen Zhou, Hao Sheng, Hailing Shi, Ling Xiao, Xiao Wang*, Jia Liu* Cell, 2023 Cover highlight in Cell Perspective highlight in Nature Methods |
(5) Autonomous experimental planning, data analysis, and scientific discovery in spatial biology with Agentic AI |
|
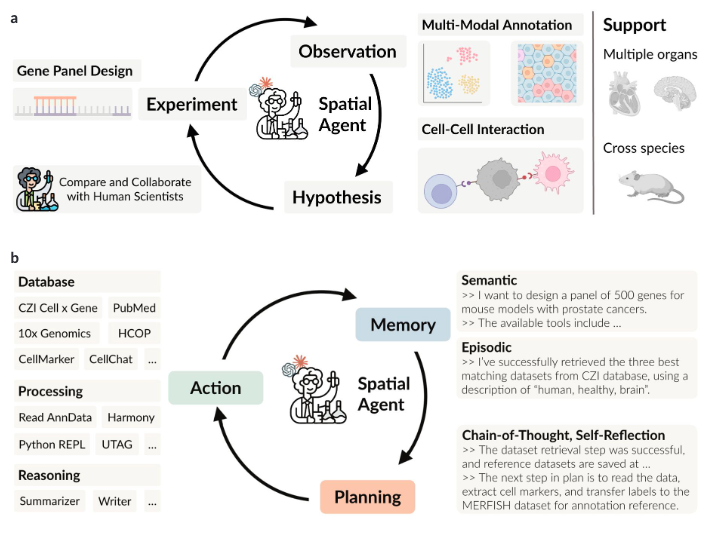
|
SpatialAgent: An Autonomous AI Agent for Spatial Biology
Hanchen Wang#*, Yichun He#, Paula Coelho#, Matthew Bucci#, Abbas Nazir, Bob Chen, Linh Trinh, Serena Zhang, Kexin Huang, Vineethkrishna Chandrasekar, Douglas C. Chung, Minsheng Hao, Ana Carolina Leote, Yongju Lee, Bo Li, Tianyu Liu, Jin Liu, Romain Lopez, Lucas Tawaun, Mingyu Ma, Nikita Makarov, Lisa McGinnis, Linna Peng, Stephen Ra, Gabriele Scalia, Avtar Singh, Liming Tao, Masatoshi Uehara, Chenyu Wang, Runmin Wei, Ryan Copping, Orit Rozenblatt-Rosen, Jure Leskovec, Aviv Regev* bioRxiv, 2025 |
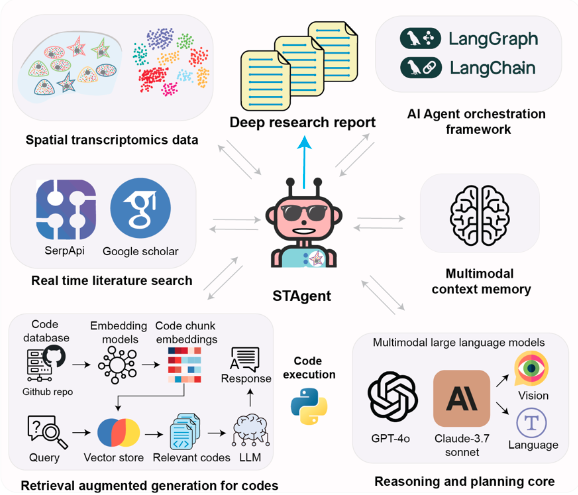
|
Spatial transcriptomics AI agent charts hPSC-pancreas maturation in vivo
Zuwan Lin#, Wenbo Wang#, Arnau Marin-Llobet, Qiang Li, Samuel D. Pollock, Xin Sui, Almir Aljovic, Jaeyong Lee, Jongmin Baek, Ningyue Liang, Xinhe Zhang, Connie Kangni Wang, Jiahao Huang, Mai Liu, Zihan Gao, Hao Sheng, Jin Du, Stephen J Lee, Brandon Wang, Yichun He, Jie Ding, Xiao Wang, Juan R Alvarez*, Jia Liu* bioRxiv, 2025 |
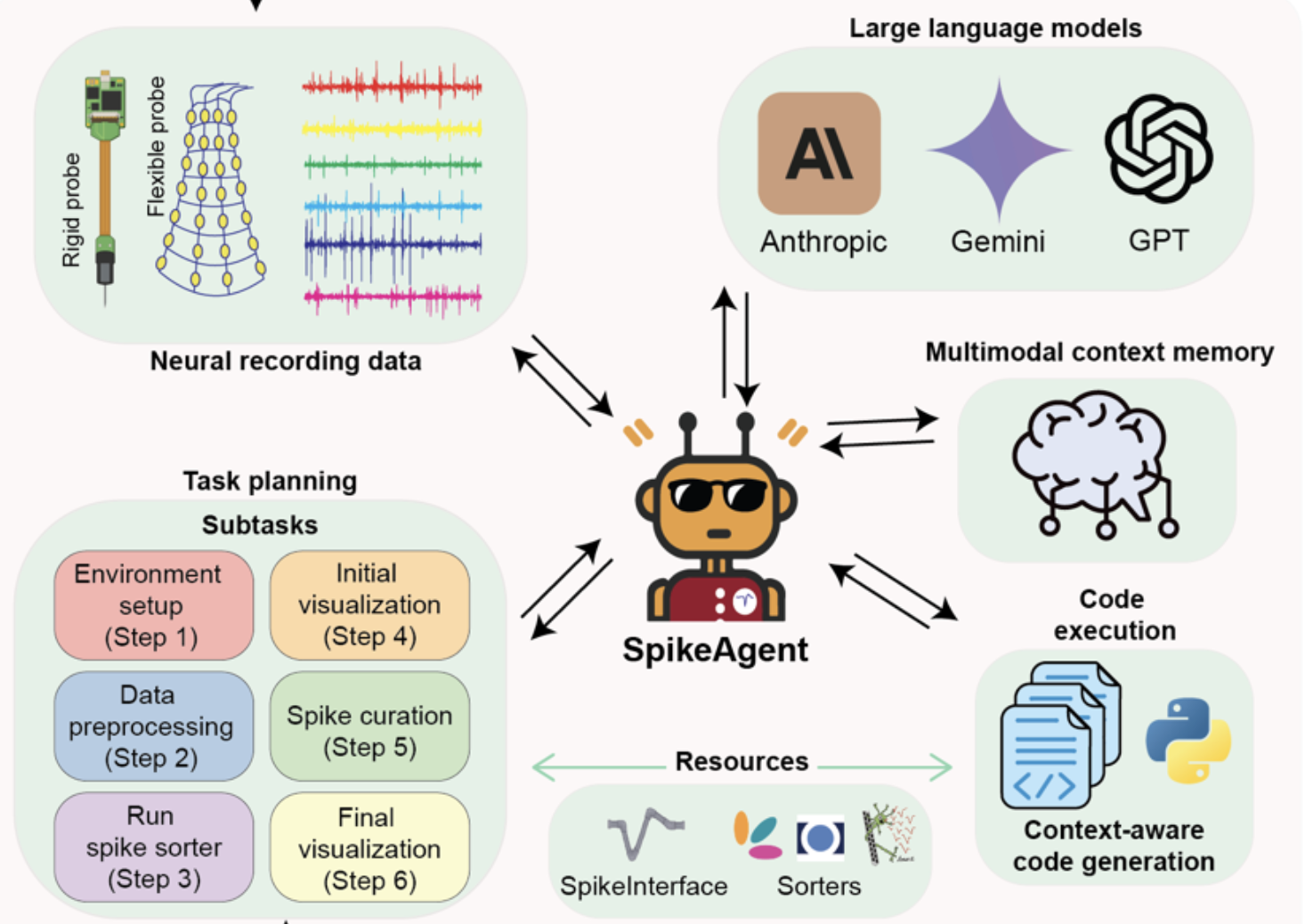
|
Spike sorting AI agent
Zuwan Lin#, Arnau Marin-Llobet#, Jongmin Baek, Yichun He, Jaeyong Lee, Wenbo Wang, Xinhe Zhang, Ariel J. Lee, Ningyue Liang, Jin Du, Jie Ding, Na Li, Jia Liu* bioRxiv, 2025 |
|
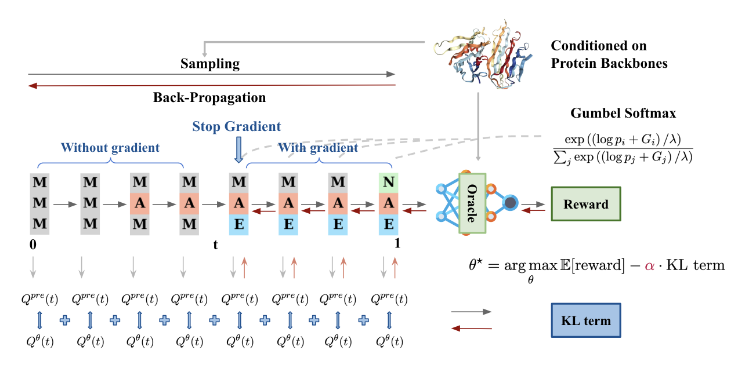
Fine-Tuning Discrete Diffusion Models via Reward Optimization with Applications to DNA and Protein Design
Chenyu Wang#, Masatoshi Uehara#, Yichun He, Amy Wang, Tommaso Biancalani, Avantika Lal, Tommi Jaakkola*, Sergey Levine*, Hanchen Wang*, Aviv Regev* NeurIPS 2024 Workshop on Machine Learning in Structural Biology, 2024 |
|
This website is borrowed from Jon Barron. Thanks! |